Chromonomer is a program designed to integrate a genome assembly with
a genetic map. Chromonomer tries very hard to identify and remove
markers that are out of order in the genetic map, when considered
against their local assembly order; and to identify scaffolds that
have been incorrectly assembled according to the genetic map, and
split those scaffolds.
Recent Changes [updated October 8, 2024]
Chromonomer Manual
What do I need to run Chromonomer?
Given a discription of an assembly, and a description of a genetic map, Chromonomer will
try to reconcile them. Chromonomer accepts standard files to perform this analysis.
- An AGP file
describing a genome assembly.
- An AGP file lays out the pattern of contigs and gaps that
make up each scaffold in an assembly. This file can be downloaded from an online
database, (e.g. Ensembl), or is produced by your assembly software.
- A tab-separated file describing all the marker positions in your genetic map.
- This file is output by genetic linkage mapping software, such as JoinMap, OneMap,
or r/QTL. Chromonomer is designed around RAD-seq markers, but any marker type that can
be aligned should work.
- A SAM or BAM file containing the alignments of markers to the assembled genome.
- The consensus sequence for each marker is aligned to the genome using any aligner.
The marker IDs for the consensus sequnce must match the marker IDs from the genetic map.
- (Optionally) A FASTA file containing the assembled genome sequence.
- If provided, a copy of the sequence will be reordered to match the chromonomed
assembly.
- (Optionally) A GFF or GTF file describing the annotation of genes in the assembled genome.
- If provided, the gene positions will be translated to match the chromonomed
assembly.
What does Chromonomer do?
Depending on the number of progeny in the genetic map (which limits the number of recombination
events), and the level of genotyping error, a genetic map will contain a certain amount of uncertainty
and error in how the markers are ordered. Similarly, depending on how a genome assembly was scaffolded,
there can be incorrect joins in the scaffolds. Chromonomer provides the best chromonome-level assembly
(a.k.a. chromonome)
given the amount of information in supplied data.
- Chromonomer will find a set of non-conflicting markers that maximizes the number of scaffolds in the
map while minimizing the nunmber of conflicted marker orders.
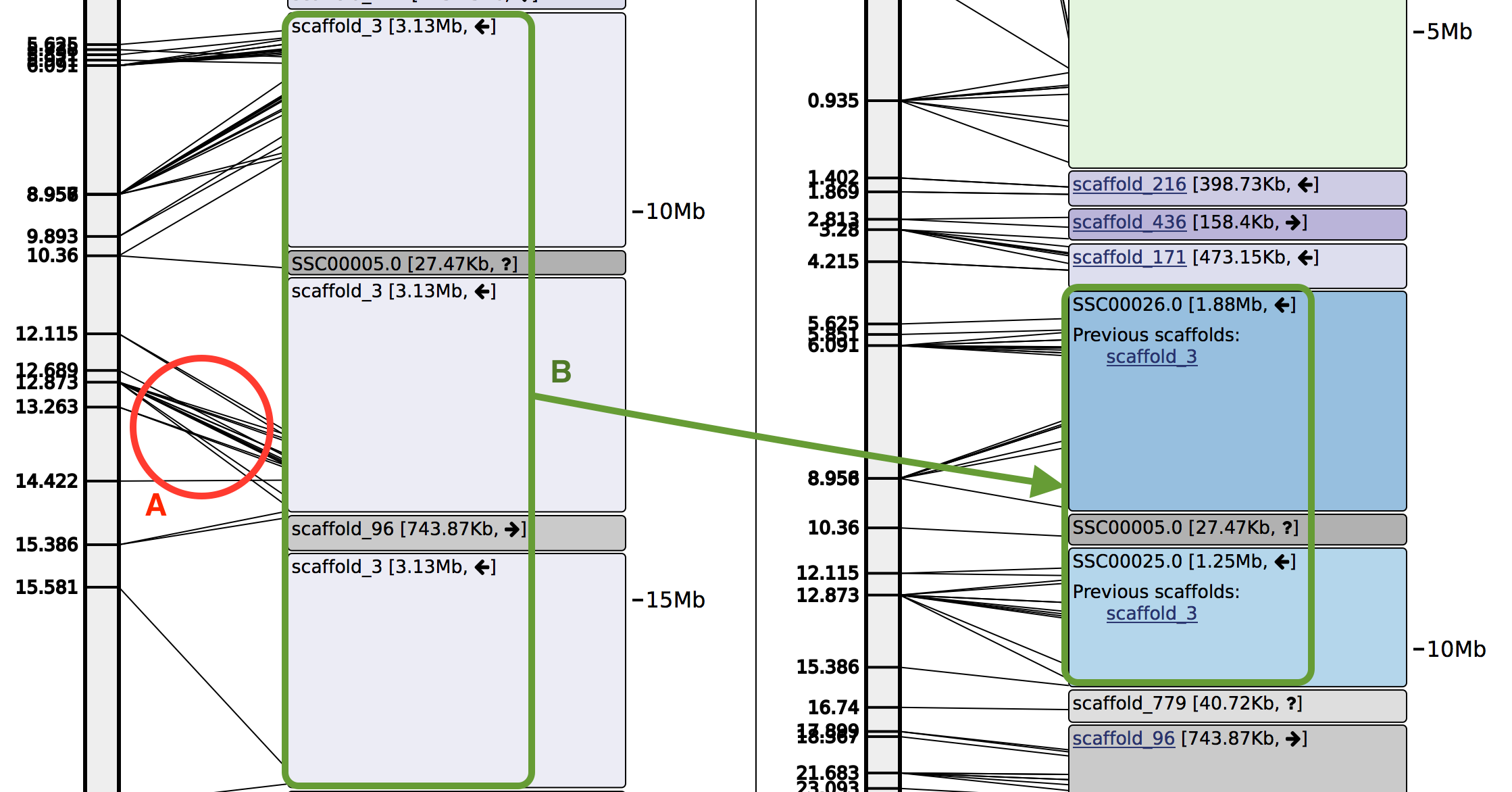
- If a scaffold spans more than one recombination point in the map, Chromonomer will assign a
forward or reverse orientation to the scaffold.
- If A scaffold occurs in more than one place along a linkage group, the scaffold will be split
at the largest gap between the two markers that mark the boundaries of the two positions. This allows
for incorrect joins to be removed and also allows for "gap-filling" by smaller scaffolds.
- Chromonomer will produce extensive documentation of this process allowing corrections to be made
in the genetic map and a method to measure different assembly iterations. If the web-interface is
installed, chromonomer will produce visualizations of each integrated linkage group.
- Chromonomer will produce new AGP and FASTA files describing the newly created chromonome.
Implementation
Chromonomer is implemented in C++. It will compile on GNU-based Linux systems
or BSD-based (Apple) OS X systems. Chromonomer is released under the
GNU GPL license.
Chromonomer was developed by Julian Catchen
<> with contributions from Angel Amores
<>.
Publications
The Chromonomer preprint describing the software is now available on bioRxiv:
- Catchen, J., Amores, A., & Bassham, S. Chromonomer: a tool set for repairing and enhancing
assembled genomes through integration of genetic maps and conserved synteny.
[reprint]
Chromonomer has been used to chromonome several genomes, including these two:
- Amores, A., Catchen, J., Nanda, I., Warren, W., Walter, R., Schartl, M., & Postlethwait, J. H.
(2014). A RAD-Tag Genetic Map for the Platyfish (Xiphophorus maculatus) Reveals Mechanisms
of Karyotype Evolution Among Teleost Fish. Genetics, 197(2), 625–641.
[reprint]
- Small, C., Bassham, S., Catchen, J., Amores, A., Fuiten, A., Brown, R., Jones, A., Cresko, W.
(2016). The genome of the Gulf pipefish enables understanding of evolutionary innovations.
Genome Biology, 17(1), 1–23.
[reprint]