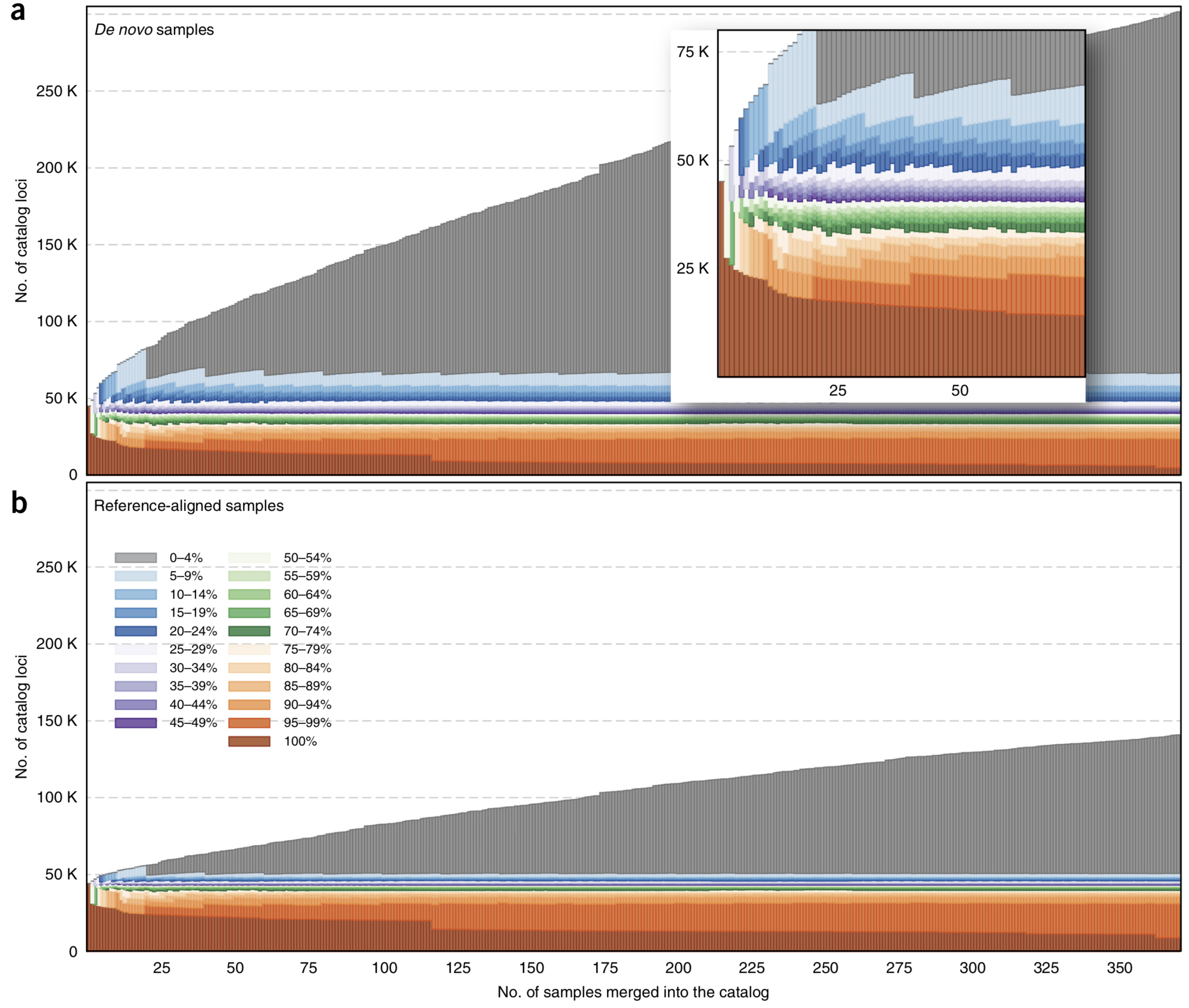
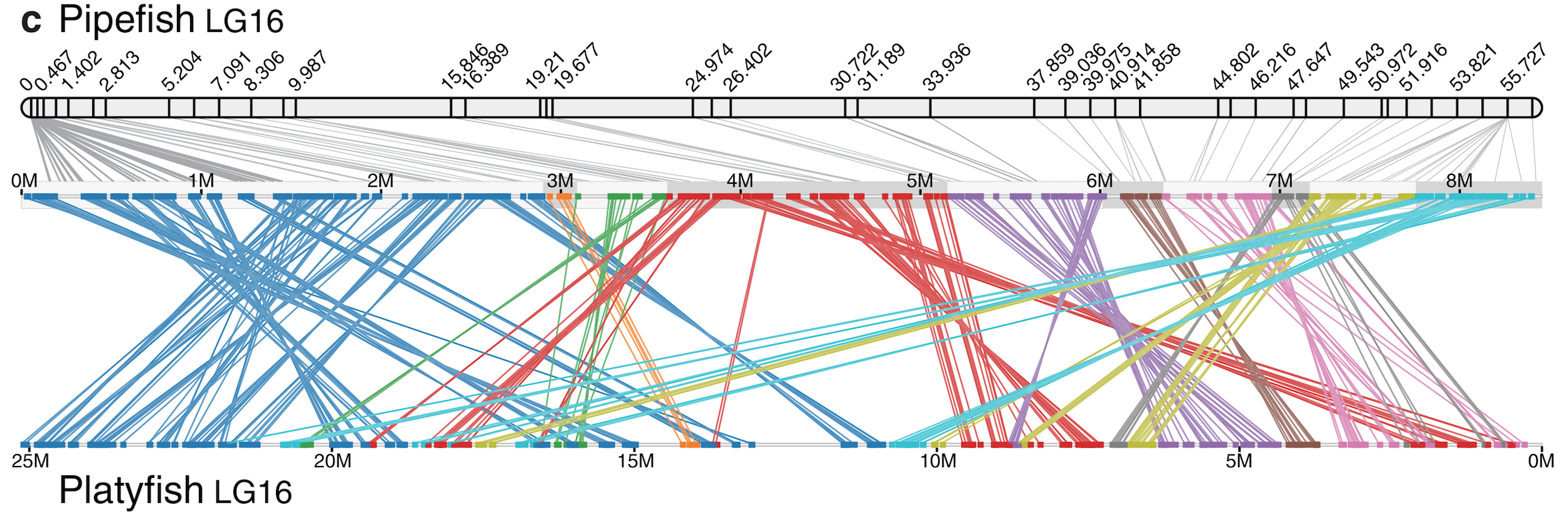
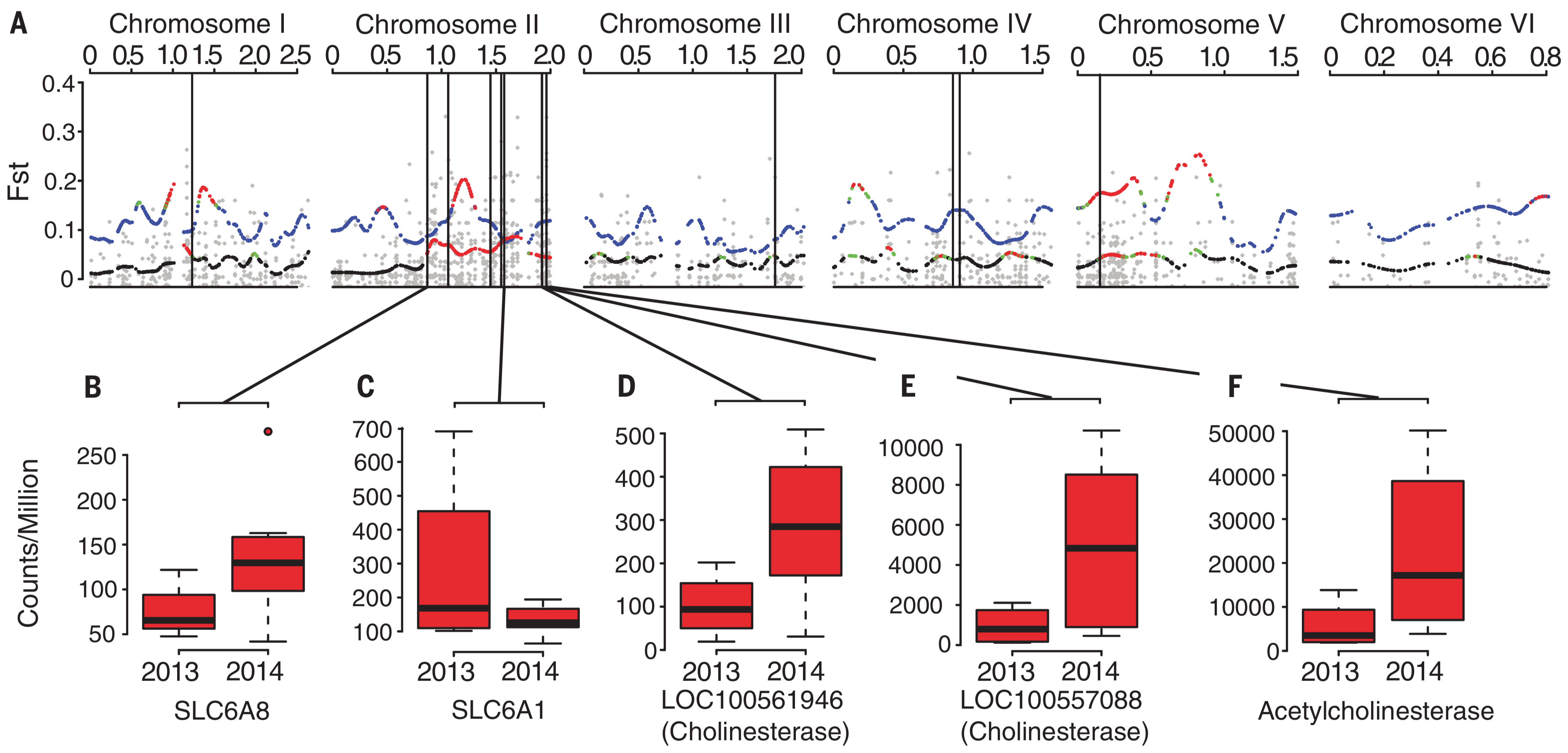
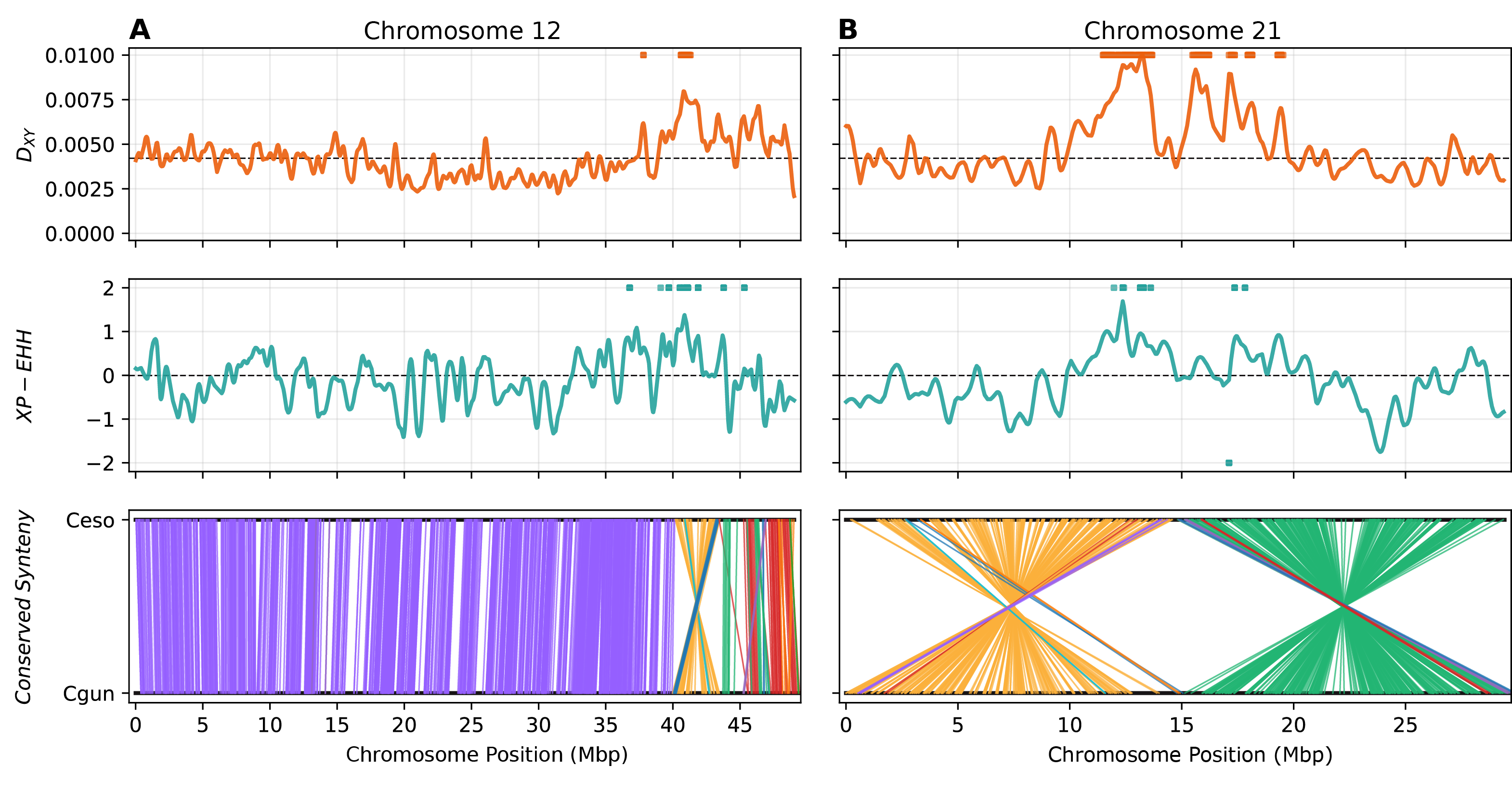
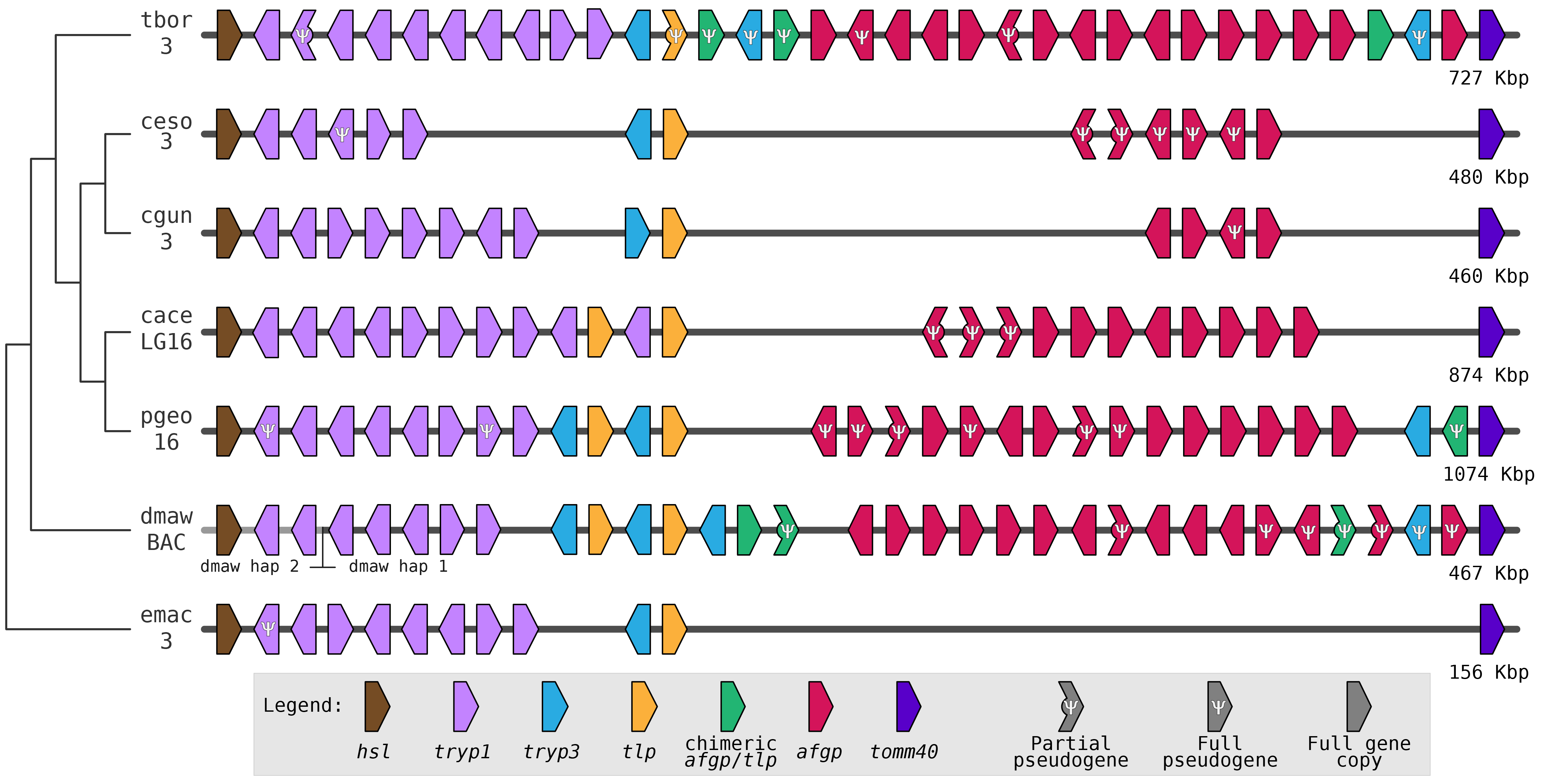
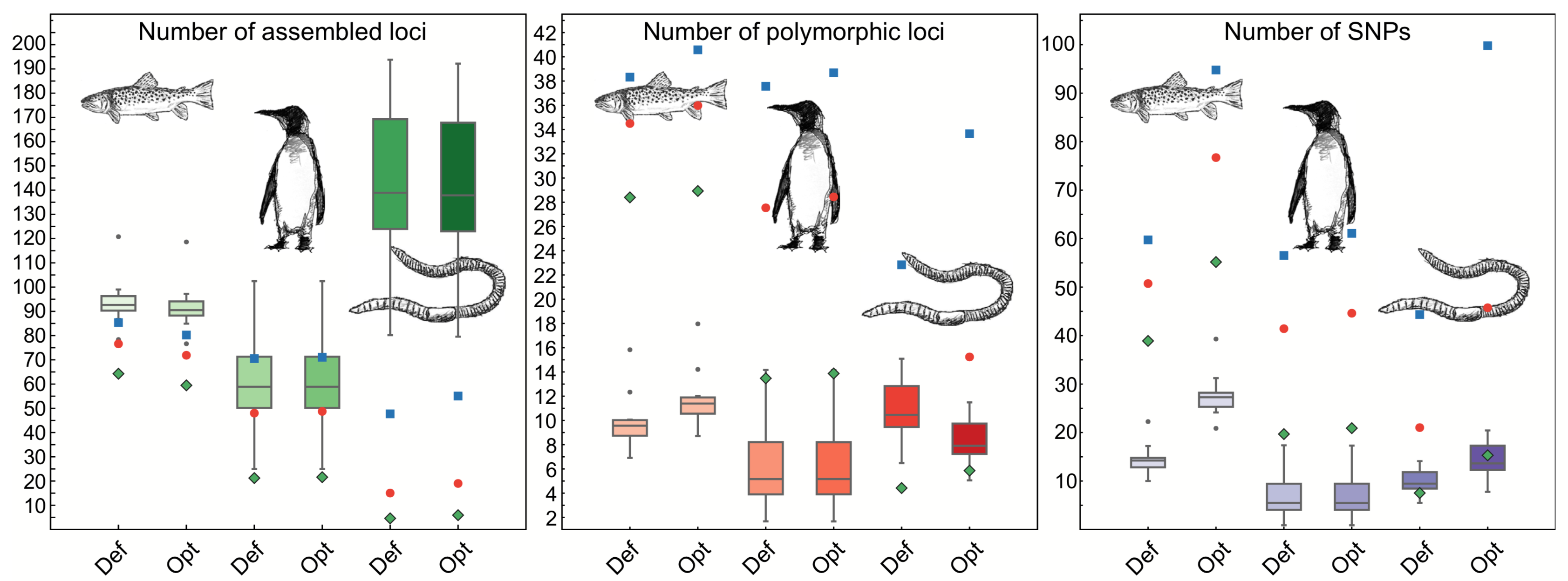






How do genomes change over evolutionary time? Numerous questions fall under this rubric, such as:
These questions can be addressed at several different scales, from whole genome duplication over millions of years to structural variation within a species over thousands of years, to somatic evolution and cancer within an individual over a handful of years. The research strategy of the Catchen Lab is to apply novel algorithmic developments to the questions of genome evolution.
The lab's work is mostly focused on teleost fish. Teleosts represent the most species rich vertebrate clade and within the teleosts lie laboratory model organisms, such as zebrafish, and several fascinating natural evolutionary models, including the threespine stickleback. We have projects focused on a number of species, from Antarctic notothenioids, to stickleback, killifish, and several modern and ancient salmon species.
 |
Jack Calvery, Graduate Student |
 |
Julian Catchen, Principal Investigator |
 |
Akane Hatsuda, Postdoctoral Research Associate |
 |
Gio Madrigal, Graduate Student |
 |
Bushra Minhas, Graduate Student |
| ⇒ Former lab members |